LATEST NEWS
Congratulations Dr. Spisak!
October 18, 2024
Congratulations to Shaun Spisak, who successfully defended his PhD thesis "High-thoughput approaches to protein engineering applied to the dCas9 and AAV2 Rep proteins."

Functional and fitness landscapes of E. coli RNase III
February 27, 2023
How protein properties such as protein activity and protein essentiality affect the distribution of fitness effects (DFE) of mutations are important questions in protein evolution. Deep mutational scanning studies typically measure the effects of a comprehensive set of mutations on either protein activity or fitness. Our understanding of the underpinnings of the DFE would be enhanced by a comprehensive study of both for the same gene. Today, Molecular Biology and Evolution published Ryan Weeks' comparative study of the fitness effects and protein activity effects of ∼4,500 missense mutations in the E. coli gene encoding RNase III. RNase III is a global regulator enzyme that cleaves diverse RNA substrates including precursor ribosomal RNA and various mRNAs. He found that although RNase III’s ability to cleave dsRNA is the most important determinant of the fitness effects of rnc mutations, the relationship between RNase III function and cell fitness was complex.Fitness was buffered to small effects on RNase III activity. Differential effects on fitness and functional scores for mutations at highly conserved residues G97, G99, and F188 suggest that these positions may be important for RNase III cleavage specificity.
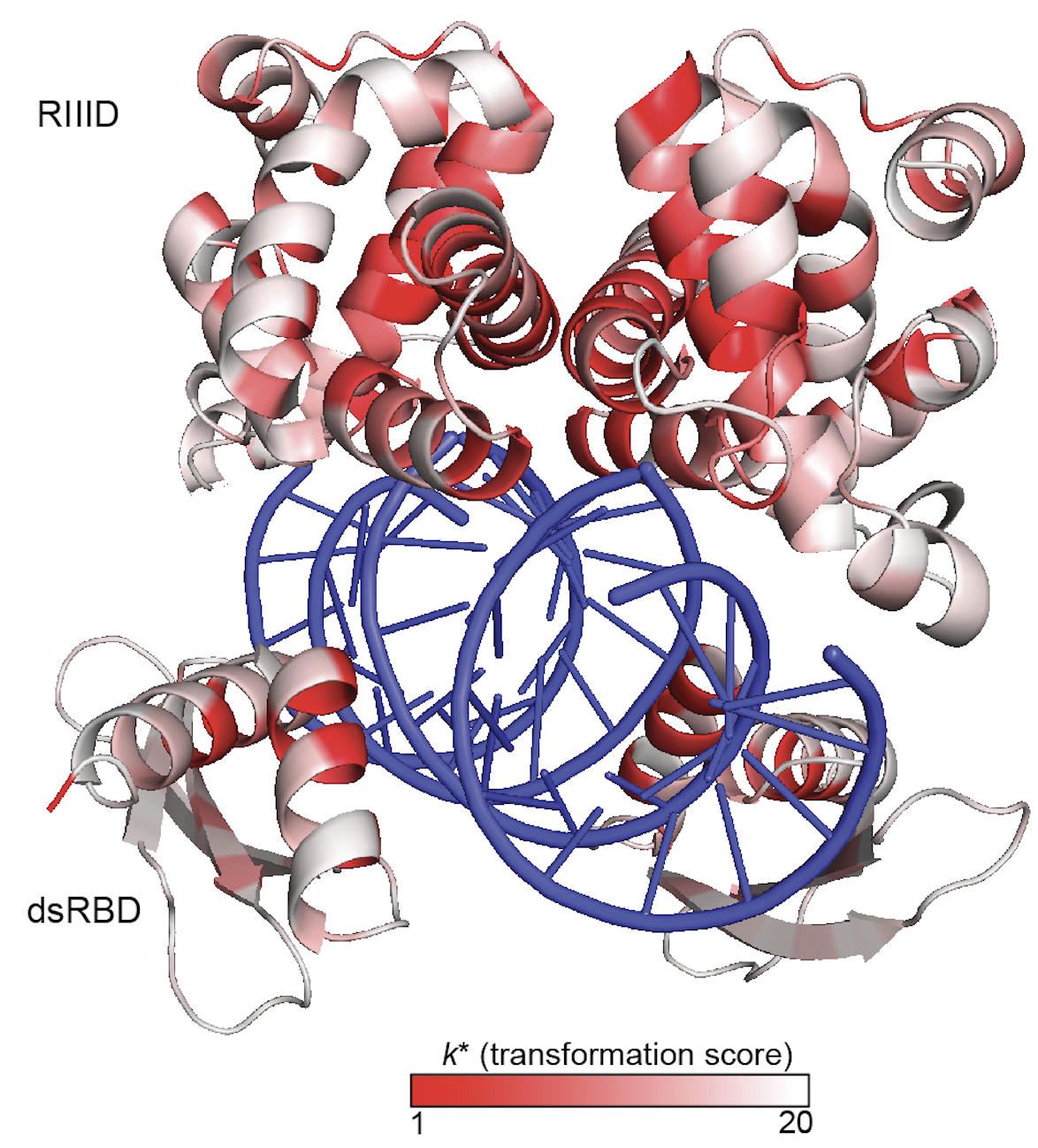
Genes vary greatly in their propensity for collateral fitness effects of mutations
February 17, 2023
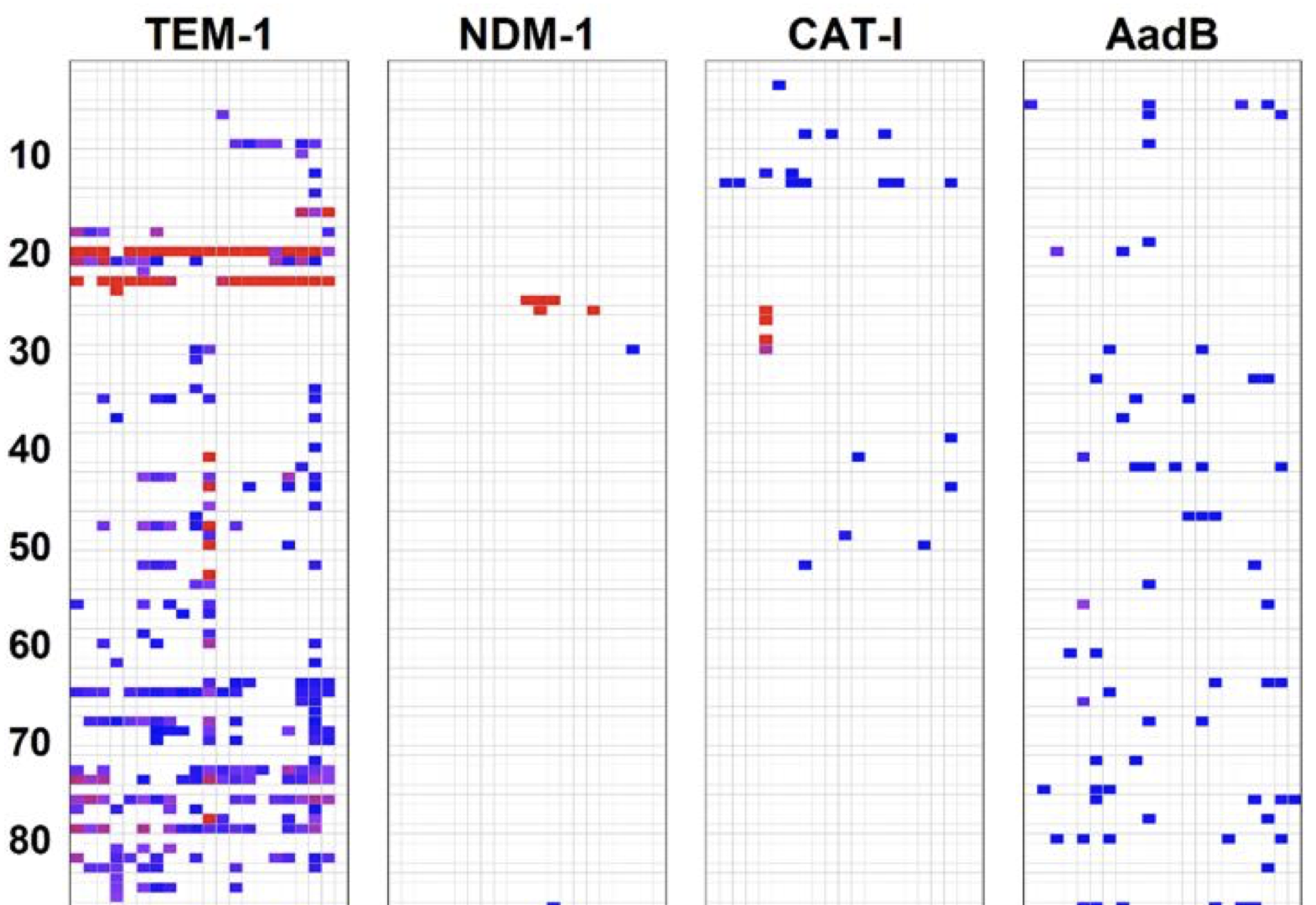
Mutations can have deleterious fitness effects when they decrease protein specific activity or decrease active protein abundance. Mutations will also be deleterious when they cause misfolding or misinteractions that are toxic to the cell (i.e., independent of whether the mutations affect specific activity and abundance). The extent to which protein evolution is shaped by these and other collateral fitness effects is unclear in part because little is known of their frequency and magnitude. Today, Molecular Biology and Evolution published Jacob Mehlhoff's systematic study of the collateral fitness effects in three antibiotic resistance genes: CAT-I, NDM-1, and aadB. He found that genes can have wildly different propensities for collateral fitness effects and that protein aggregation did not correlate with these effects for many of the deleterious mutants in CAT-I and NDM-1. Select deleterious mutantations caused unexpected phenotypes to emerge. The introduction of internal start codons in CAT-1 caused loss of the episome and, even stranger, a mutation in aadB made its cognate antibiotic essential for growth. Jacob's study illustrates how the complexity of the cell provides a rich environment for collateral fitness effects and new phenotypes to emerge.
A dCas9 system for assaying and selecting RNase III activity
December 9, 2022
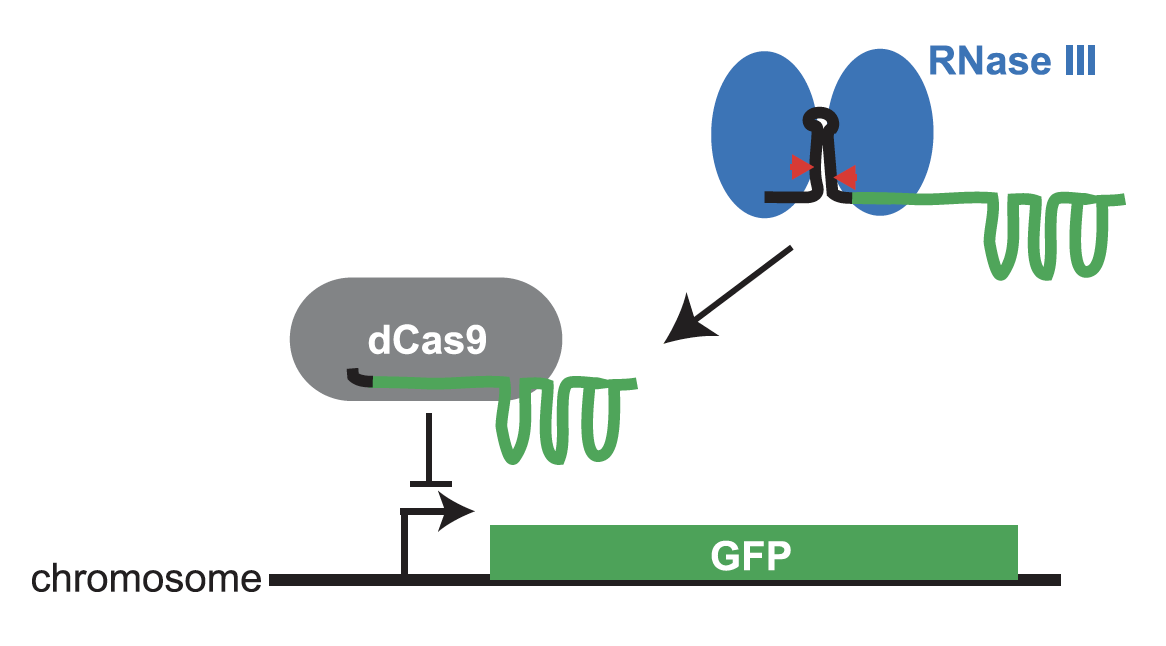
Pricila Hauk and Ryan Weeks have developed a CRISPR/dCas9 system to assay and select for the ribonuclease activity of RNase III in E. coli. This work was published today in CRISPR J. The key molecule in the system is a hybrid guide RNA (gRNA) between an RNase III substrate and a gRNA that works with dCas9 to repress GFP expression. This fusion must be cleaved by RNase III for full GFP repression. Our system uses GFP fluorescence to report on Escherichia coli RNase III activity in culture and on an individual cell basis, making it effective for selecting individual cells through fluorescence-activated cell sorting. In a forthcoming paper, Ryan Weeks has used the system to determine the in vivo functional effects of thousands of mutations in E. coli RNase III.
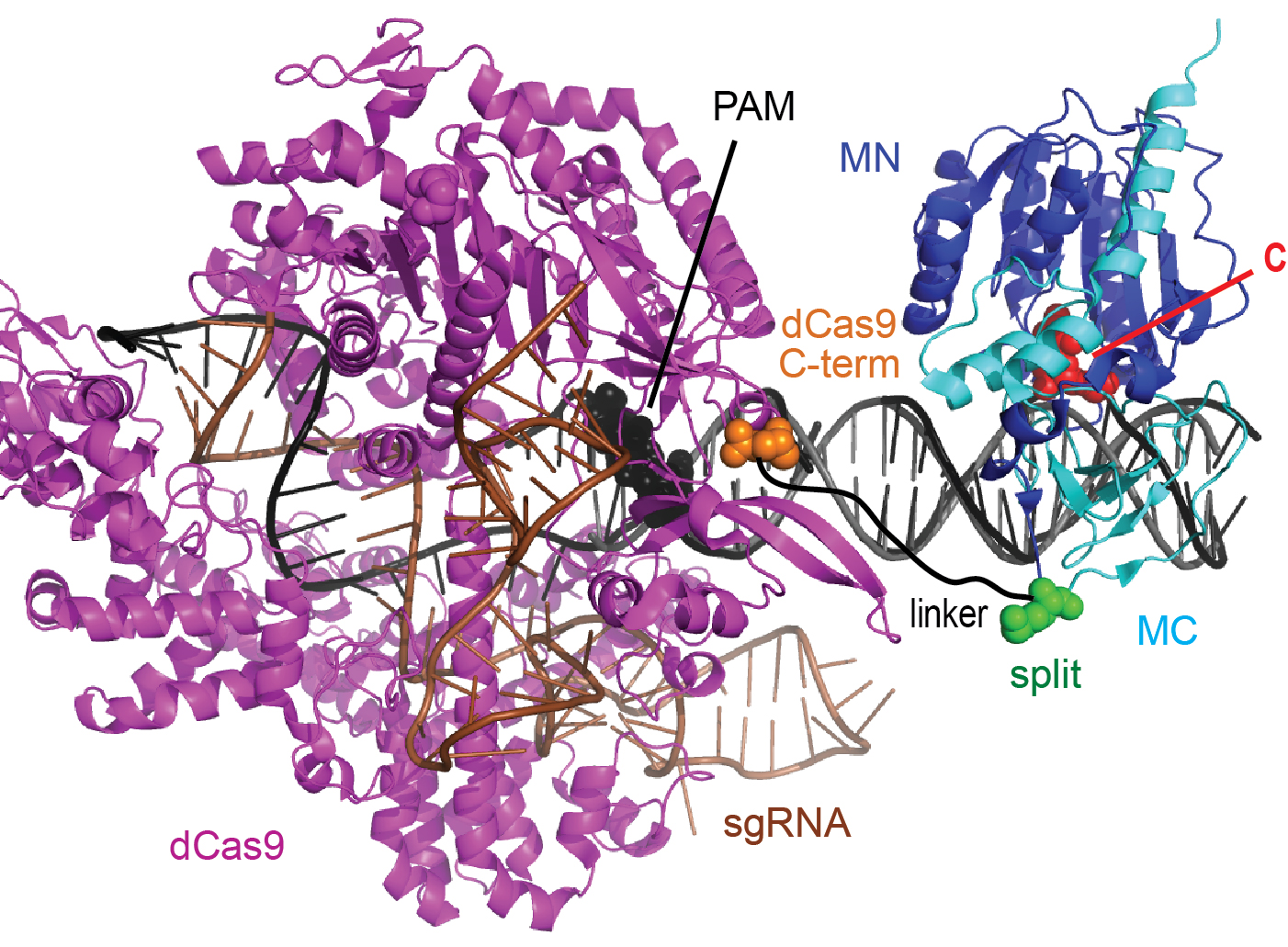
A dual selection system for dCas9 activity
June 3, 2022
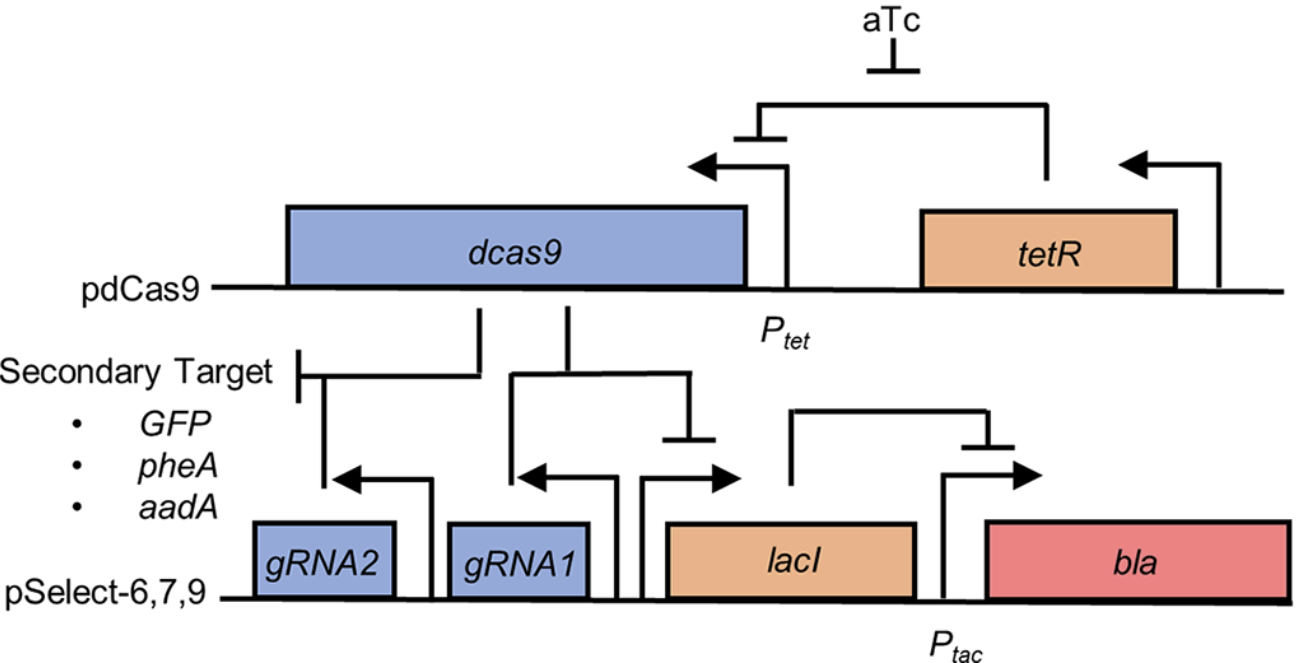
The nuclease-null CRISPR protein dCas9 (aka CRISPRi) can repress gene expression at sites determined by its cognate guide RNA. The technology would benefit from ways to control this repression exogenously using molecular signals. The creation of such dCas9 switch proteins would benefit from a single system for both positive and negative selection of dCas9 activity. Shaun Spisak has developed such a system, and a paper describing this work was published today in PLOS ONE. This system can be used in E. coli to perform both positive and negative selections for dCas9 activity without additional cloning or transformation steps between selections.
Congratulations Dr. Weeks!
May 20, 2022
Ryan Weeks successfully defended his PhD thesis "Fitness and Functional Landscapes of RNase III."
Nisita Dutta and Olek Pisera receive graduate fellowships
April 13, 2022
Two former Ostermeier lab students recently received prestigious graduate fellowships.
Nisita Dutta (MSE ChemBE 2020) was recently awarded a Gates Cambridge Scholarship, which provides her full funding to pursue a PhD in Chemistry at the University of Cambridge. Nisita will be working with doctors to create nanobody-drug conjugates that target mesothelin, a cell-surface protein expressed on pancreatic cancer cells.
Olek Pisera (BS ChemBE 2017) has been named a 2022 Paul & Daisy Soros Fellow. This fellowship will support his PhD studies in Biomedical Engineering at the University of California, Irvine where he is developing a synthetic biology system that could allow for discovery of antibodies against classes of mammalian cell surface proteins that have been inaccessible with current technologies.
Congratulations Dr. Mehlhoff!
March 14, 2022
Jacob Mehlhoff successfully defended his PhD thesis "Collateral fitness effects of mutations."
Marc Ostermeier elected AAAS Fellow
January 26, 2022
Marc Ostermeier was elected a Fellow of the American Association for the Advancement of Science in the Biological Sciences Section. Read the HUB article on this year's Hopkins' inductees.
New NSF grant to study collateral fitness effects
April 20, 2021
The Ostermeier lab received a ~$1 million grant from NSF-MCB from the Genetic Mechanisms Program to continue our study of collateral fitness effects.
Ostermeier lab certified green thanks to the efforts of Ryan Weeks
February 4, 2021
In December the Ostermeier Lab completed the My Green Lab green lab certification. The program focuses on the behavioral changes that labs can make to reduce their impact on the environment such as setting the -80°C freezer at -70°C, turning off equipment at night, reducing waste, and recycling. The certification represents the global standard for sustainable lab behaviors recognized by the Association for the Advancement of Sustainability in Higher Education, the American Energy Society, and the International Institute for Sustainable Laboratories. Our lab achieved the highest level of certification, green, with over 80% of best lab practices implemented.
The effort was led by PhD student Ryan Weeks, who has also helped launch a pilot program for the university to help 25 Hopkins labs through the certification process as described in a recent HUB article.

Jacob Mehlhoff and Ryan Weeks helping to make N95 masks
May 26, 2020
During the cornovirus research shut down at Hopkins, Jacob Mehlhoff and Ryan Weeks have been volunteering to help make filters for N95 masks at DiPole Materials, an electrospinning company in the Baltimore neighborhood of Pigtown. In total, 13 current students and graduates from the Whiting School of Engineering and Krieger School of Arts and Sciences are now working on the project. By operating 24 hours a day with the help of Hopkins students, the company makes enough material for about 2,000 mask filters per day.
See the story on HUB and the video below.
Jacob Mehlhoff receives the WSE Outstanding TA Award
May 13, 2020
Jacob Mehlhoff received the Whiting School of Engineering Award for Outstanding Teaching Assistants for his performance as a TA in Process Design with Aspen.
Congratulations Nisita!
May 9, 2020
Nisita Dutta successfully presented her masters essay "Characterizating the effects of the Psp and Rcs stress pathways on collateral fitness effects in E. coli" This fall Nisita will be starting her MD/PhD with the University of Maryland Medical Scientist Training Program and the NIH Oxford/Cambridge Scholars Program.
Collateral fitness effects of mutations
May 9, 2020
Today, PNAS published our latest work studying the effects of mutations using deep mutational scanning. Mutations provide the source of genetic variability upon which evolution acts. Deleterious protein mutations are commonly thought of in terms of how they compromise the protein’s ability to perform its physiological function. However, mutations might also be deleterious if they cause negative effects on one of the countless other cellular processes. The frequency and magnitude of such collateral fitness effects are unknown. Our systematic study of mutations in a bacterial protein finds widespread collateral fitness effects that were associated with protein aggregation, improper protein processing, incomplete protein transport across membranes, incorrect disulfide-bond formation, induction of stress-response pathways, and unexpected changes in cell properties. The surprising prevalence of deleterious collateral fitness effects suggests they may play a role in constraining protein evolution.
The multi-year effort was led by Jacob Mehlhoff and Frank Stearns with contributions by current and former lab members Dahlia Rohm, Jerry Wang, Henry Tsou, Nisita Dutta, Erin Hsiao, and Courtney Gonzalez with assistance from Alan Rubin at the Walter and Eliza Hall Institute of Medical Research.
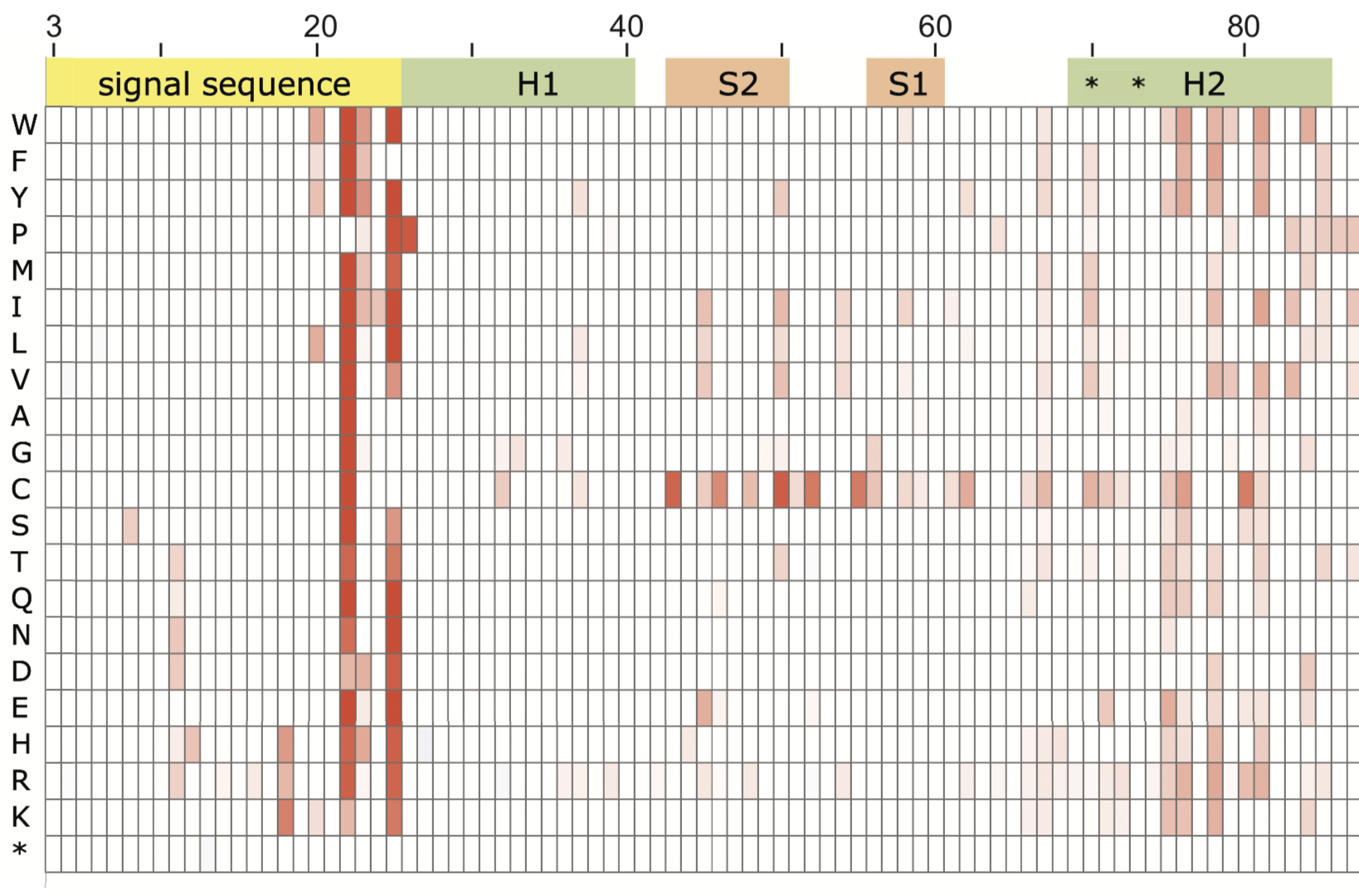
Congratulations Jerry!
July 11, 2019
Buheng (Jerry) Wang successfully presented his masters essay "Collateral fitness effects of signal peptide mutants of beta-lactamase."
New proteins for gene-directed enzyme prodrug therapy
June 6, 2019
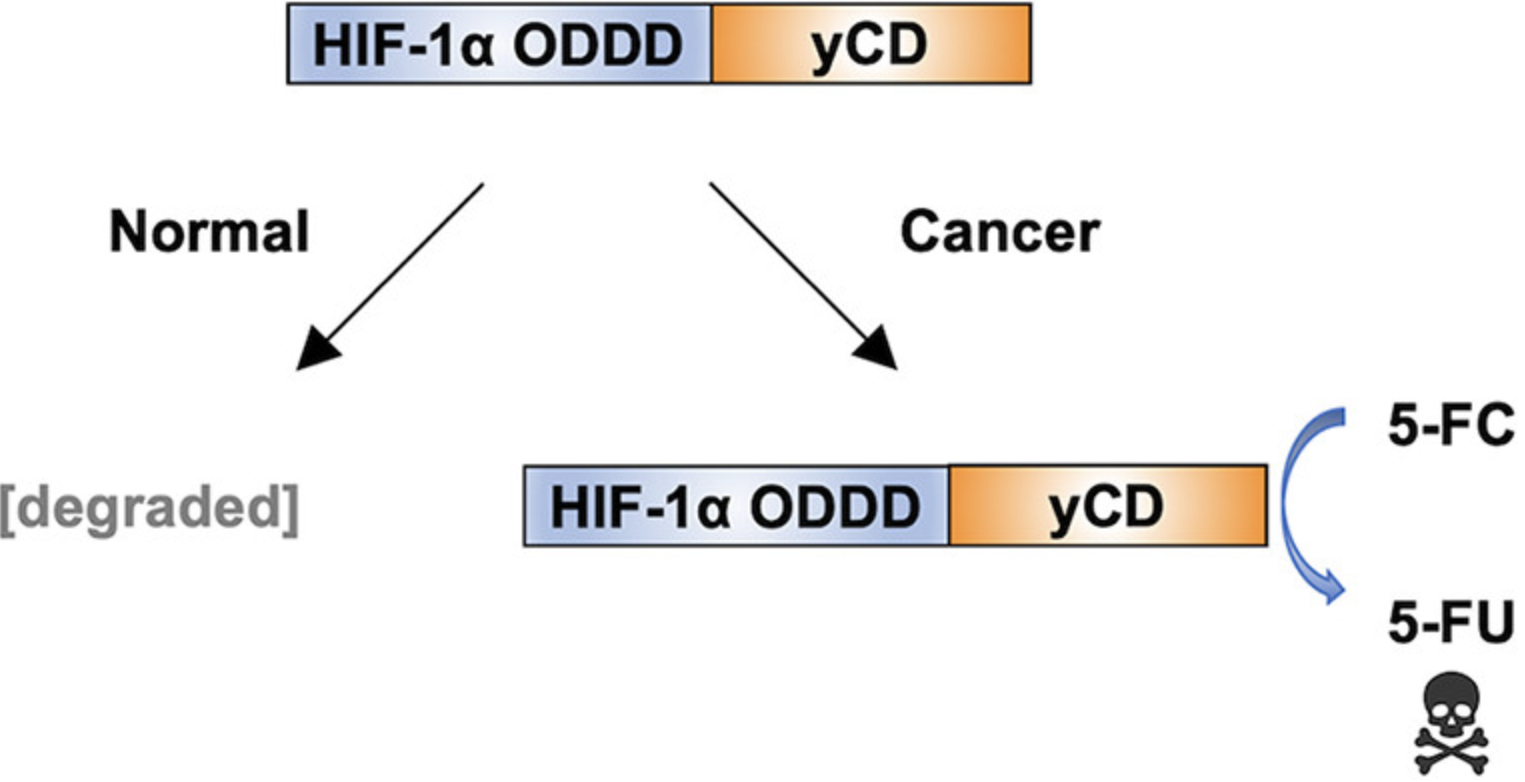
Two papers were recently published from Tiana Warren's thesis work. Both of the papers involve the yeast enzyme cytosine deaminase (yCD), which together with the prodrug 5-fluorocytosine (5FC) has potential to be used therapeutically in a strategy called Gene-Directed Enzyme Prodrug Therapy (GDEPT). In this strategy, the gene encoding yCD is delivered selectively to cancer cells. Upon subsequent administration of 5FC, the yCD in cancer cells selectively converts 5FC to 5-fluorouracil, a chemotherapeutic. Two limitations of current GDEPT technology are difficulty in selectively delivering the yCD gene to cancer cells and lower than optimal yCD activity in cancer cells.
In a paper appearing in ACS Synthetic Biology, Tiana and her coauthors developed a version of yCD that is selectively degraded in cancer cells because it is fused to a protein domain that is targeted for degradation in hypoxic cells, such as those in tumors. These HOPE fusions have the potential to address the selectivity issue of gene delivery. The second paper appears in AIChE Journal. Tiana and her coworkers used comprehensive site saturation mutagenesis and directed evolution to identify yCD mutations that improve its ability to kill cells in the presence of 5FC. This paper will appear in a special Tribute to Founders issue of AIChE Journal honoring Nobel Laureate Frances Arnold, who was awarded the 2018 Nobel Prize in Chemistry for pioneering the directed evolution of enzymes.
Coauthors of the papers include ChemBE undergraduates Krishna Patel and Jordan Rivera and our JHMI collaborator James Eshleman.
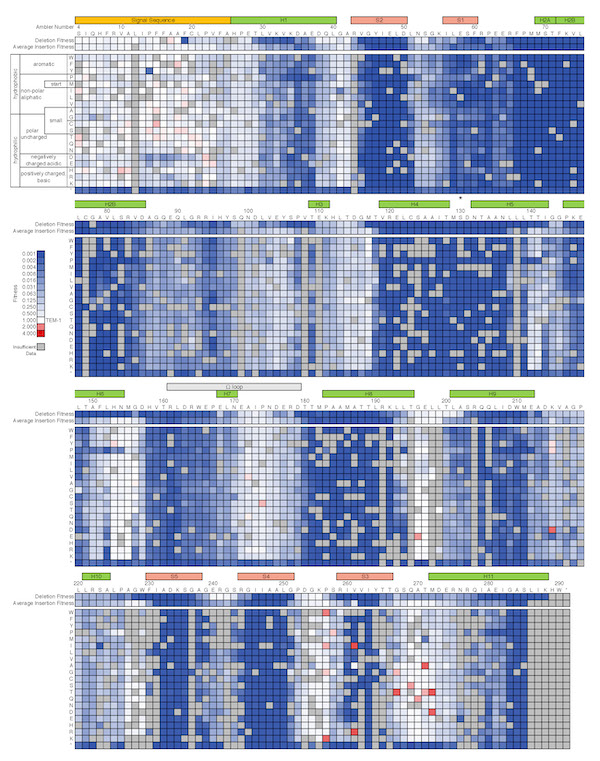
Fitness effects of InDels and sequential mutations in TEM-1
May 9, 2019
The Journal of Molecular Biology recently published two papers by Courtney Gonzalez in which she mapped regions of TEM-1 β-lactamase's fitness landscape using deep mutational scanning
Short insertions and deletions (InDels) are a common type of mutation found in nature and a useful source of variation in protein engineering. InDel events have important consequences in protein evolution, often opening new pathways for adaptation. Yet much less is known about the effects of InDels compared to point mutations and amino acid substitutions. In particular, deep mutational scanning studies on the distribution of fitness effects of mutations have focused almost exclusively on amino acid substitutions. In Courtney's first paper (coauthored by former ChemBE undergradaute student Paul Roberts) she determined the effect of 78% of the 5720 possible amino acid insertions and 98% of the 286 possible amino acid deletions in TEM-1 β-lactamase. This first near-comprehensive examination of the fitness effects of InDels revealed partially overlapping deletion-tolerant and insertion-tolerant regions throughout the protein, especially in unstructured regions and at the end of helices.
In Courtney's second paper she quantified the extent of epistasis between ~8,000 pairs of sequential mutations and found that sequential mutations are particularly prone to interact negatively.
Congratulations Gareth!
April 29, 2019
Gareth Evans successfully presented his masters essay "Creation of modular inducible respressors via directed evolution."
Improved targeted DNA methyltransferases
December 18, 2018
Our latest paper on targeted DNA methylation using CRISPR/dCas9 appeared today in PLoS ONE . We demonstrate how linker length and mutations that reduce the methyltransferase's affinity for DNA can improve targeting of DNA methylation to desired CpG sites. Many thanks to our collaborators in the labs of Winston Timp (Hopkins BME) and Carl Novina (Dana-Farber Cancer Institute).
Congratulations Dr. Gonzalez!
October 5th, 2018
Courtney Gonzalez successfully defended her thesis "Exploring the fitness landscape of TEM-1 β-lactamase: a survey of intragenic epistasis and the fitness effects of InDels."
Congratulations Dr. Warren!
September 28th, 2018
Tiana Warren successfully defended her thesis "Protein engineering improvements to enzyme/prodrug therapy using yeast cytosine deaminase."
Two new NSF grants
June 28, 2018
The Ostermeier lab received two new NSF grants. The aim of the first (from NSF-CBET) is to develop modular protein switches. The aim of the second (from NSF-MCB) is to study underexplored mechanisms that constrain the evolution of proteins.
Congratulations Dr. Xiong!
November 2nd, 2017
Earlier this semester, Tina Xiong successfully defended her thesis on using CRISPR/Cas9 to target DNA methylation of CpG sites. A large portion of her thesis work was published in July in Scientific Reports, with a follow up paper soon to be submitted. Tina is now off to a Senior Scientist position at Merck.
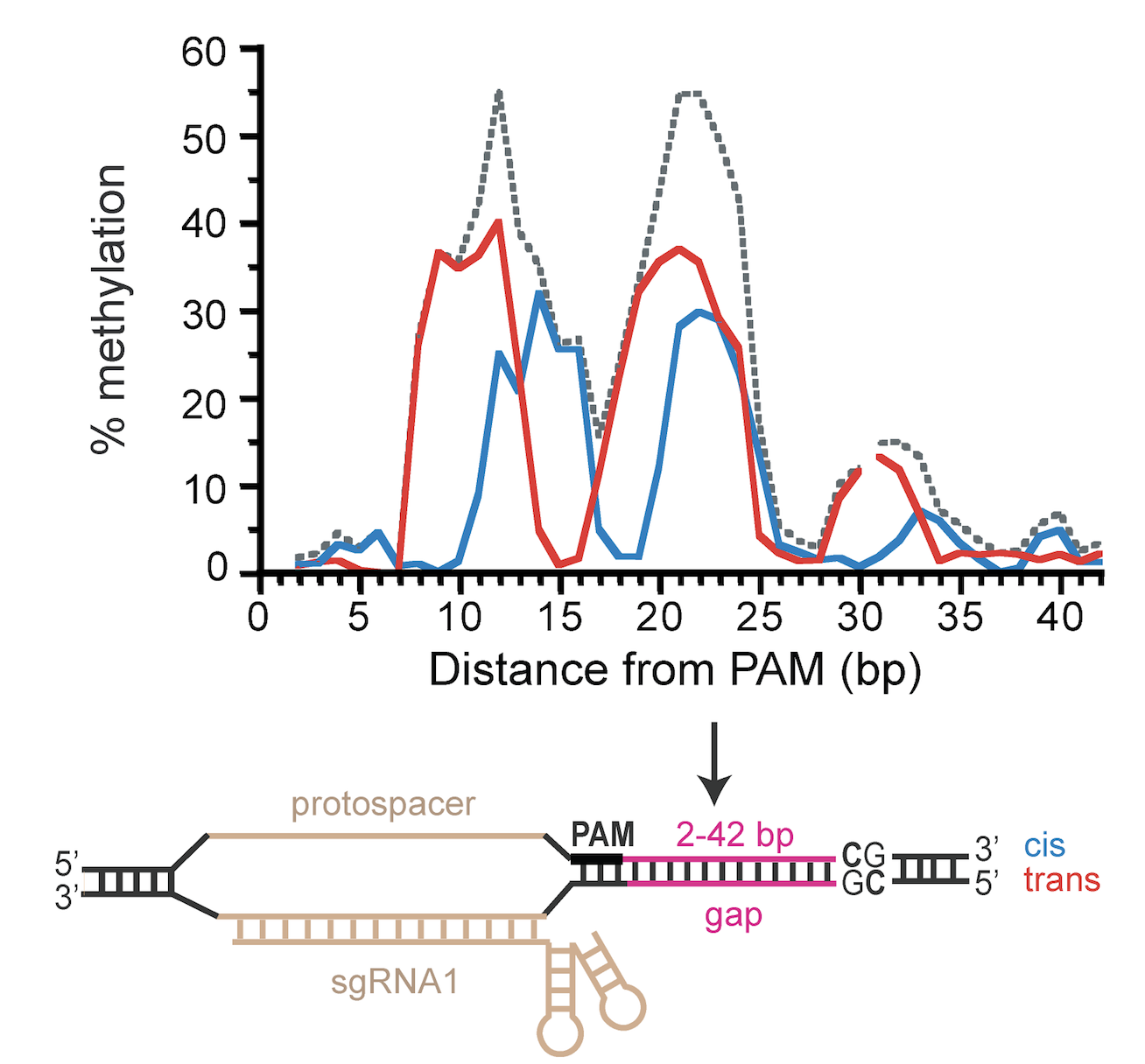
RNA-programmed DNA methylation using CRISPR/Cas9
July 27, 2017
Mammalian genomes exhibit complex patterns of gene expression regulated, in part, by the level of DNA methylation. The advent of engineered DNA methyltransferases (MTases) to target DNA methylation to specific sites in the genome will accelerate many areas of biological research. However, targeted MTases require clear design rules to direct site-specific DNA methylation and minimize the unintended effects of off-target DNA methylation.
Today, Scientific Reports published Tina Xiong's paper describing our use of CRISPR/Cas9 to target methylation. This work is a collaboration between our lab, the Winston Timp lab (JHU), and the Carl Novina lab (Dana-Farber Cancer Institute) including Ostermeier lab alumn Glenna Meister. We describe a targeted MTase composed of an artificially split CpG MTase (sMTase) with one fragment fused to a catalytically-inactive Cas9 (dCas9) that directs the functional assembly of sMTase fragments at the targeted CpG site based on guide RNA sequences. We precisely map RNA-programmed DNA methylation to targeted CpG sites as a function of distance and orientation from the protospacer adjacent motif (PAM). Expression of the dCas9-sMTase in mammalian cells led to predictable and efficient (up to ~70%) DNA methylation at targeted sites. Multiplexing guide RNAs enabled targeting methylation to multiple sites in a single promoter and to multiple sites in multiple promoters. This programmable de novo MTase tool might be used for studying mechanisms of initiation, spreading and inheritance of DNA methylation, and for therapeutic gene silencing.
Tiana Warren receives the 2017 JHU Diversity Recognition Award
May 18, 2017
Congratulations to Tiana Warren for receiving a 2017 JHU Diversity Recognition Award! Tiana has had leadership roles in several organization with the Whiting School of Engineering, most notably the Black Graduate Student Association, the National Organization for the Professional Advancement of Black Chemists and Chemical Engineers, and the Johns Hopkins Diversity Leadership Council. As a KSAS/WSE Graduate Diversity Fellow she has advised divisional leadership on issues related to graduate student diversity and inspired the production of a Homewood-based, graduate URM resource and community handbook.

Elad Firnberg on NPR's Marketplace talking about Revolve
September 30th, 2016
NPR's Marketplace talked to Ostermeier lab alumnus Elad Firnberg yesterday about how free undergrad tuition at Cooper Union enabled him to co-found Revolve Biotechnologies. Revolve Biotechnologies is a life science company focused on engineering better proteins using novel directed evolution technologies. The company's IP portfolio was developed in our lab. In addition to its internal protein pipeline, Revolve is currently offering a service for construction of custom DNA variant libraries.

Congratulations Dr. Shelat!
September 27th, 2016
Earlier this month, Nirav Shelat successfully defended his thesis on engineering the herpes simplex virus thymidine kinase for enzyme prodrug therapy. PLos ONE just published his paper on the development of a new positive genetic selection for nucleoside kinase activity in E. coli.
Tina Xiong wins poster award at SEED!
July 29th, 2016
Congratulations to Tina Xiong for winning a Biotechnology Journal-sponsored poster award at the 2016 Synthetic Biology: Engineering, Evolution & Design (SEED) conference in Chicago last week for her poster entitled "RNA-programmed DNA methylation". Tina earned one of only six poster awards, and there were 234 posters!

Survey of epistasis along an adaptive pathway
June 10th, 2016
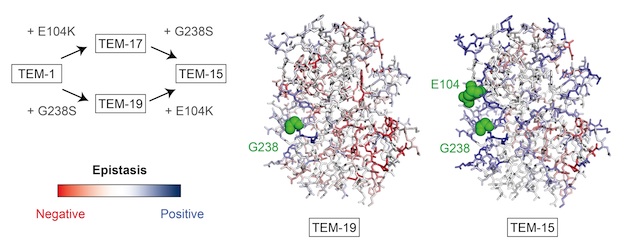
An understanding of the role of intragenic epistasis and tradeoffs in adaptation is central to an understanding of protein evolution. Today, Journal of Molecular Biology published Barrett's extensive and systematic study of a series of fitness and epistatic landscapes along an adaptive pathway (Shifting fitness and epistatic landscapes reflect trade-offs along an evolutionary pathway J. Mol. Biol. 428, 2730–2743), the first study of its kind. He measured the effect on ampicillin resistance of ~12,500 unique mutants of alleles of TEM-1 β-lactamase along an adaptive path in the evolution of cefotaxime resistance. This series of fitness and epistatic landscapes provided extensive experimental insight into the relationships between mutation, protein structure, protein stability, and epistasis and revealed the tradeoffs inherent in the evolution of new functions. We found pervasive epistasis involving adaptive mutations that can be partially understood in terms of protein structural and stability considerations. Adaptation moved the protein to a more rugged and precarious region of the fitness landscape, which is a cost of adaptation.
When bad is good
January 29, 2016
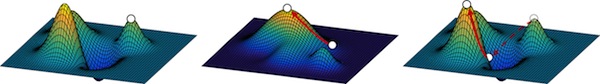
Barrett's paper Environmental changes bridge evolutionary valleys (Sci. Adv. 2(1):e1500921) was highlighted in "This Week In Science" in today's issue of Science. The paper demonstrates that evolution through inferior intermediates can paradoxicaly lead to superior outcomes.
Barrett subjected TEM-15 beta-lactamase to four different selection regimen, three of which involved environmental changes that alter the fitness landscape. The "negative selection" regimen (which included steps for selection of mutations that substantially decrease antibiotic resistance) outperformed the other strategies and ultimately resulted in proteins conferring the highest resistance. In the analysis of one such high resistance protein, he found that an initial, severely deleterious mutation is an initial gateway to a relatively inaccessible area of sequence space. Furthermore, this mutation participates in higher-order positive epistasis with a number of beneficial mutations, compensating for the increasing negative epistasis between beneficial mutations as they accumulate (i.e. compensating for diminishing returns). The ability of "negative" selection and environmental changes to provide access to novel fitness peaks has important implications for natural evolutionary mechanisms as well as applied directed evolution.
Modular protein switches built from antibody mimetics
December 7, 2015
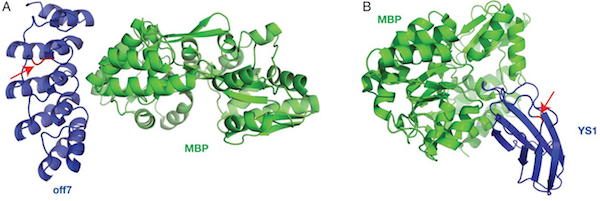
In a just published paper in Protein Engineering Design and Selection, we demonstrate that protein switches can be built using DARPins and monobodies as input domains. The DTRA and NIH funded research was initiated by Manu Kanwar and Amol Date and fully realized by first author Nate Nicholes (with some assistance from Pamphile Beaujean and Pricila Hauk).
Our manuscript describes how domain insertion can be used to create protein switches that consist of fusions between antibody mimetic proteins (monobodies and DARPins) and the enzyme beta-lactamase. These switches' enzyme activity is modulated by the antibody mimetic domain’s ligand. We demonstrate that these switches exhibit modularity in the sense that new switches can be created simply through the introduction of mutations in the antibody mimetic domain that are known to cause binding to the new ligand. There are two significant conclusions from our work. First, it is possible to create protein switches by domain insertion using input domains that do not undergo large conformational changes upon ligand binding. Second, DAPRins and, to a lesser extent, monobodies show promise as input domains for a modular platform for the rapid development of switches.
Two recent protein switch papers
October 13, 2015
Two of our papers on protein switches have recently appeared in Biotechnology and Bioengineering.
In the first paper, Jay Choi together with Maya Zayats from Peter Searson's lab show how an electrochemical signal can be used as an exogenous input to control protein switch function via reduction of the engineered disulfide bonds. This study suggests that a disulfide-containing protein switch is a potentially useful platform for bioelectronic sensors with remote control of the sensing ability.
In the second paper, Nate Nicholes and Jen Tullman led an effort to create switches by fusing TEM-1 beta-lactamase and a variety of paralogous periplasmic binding proteins. The results indicate that the emergence of the switch property likely depends on the precise molecular details of the fusions and cannot be easily predicted from some overall general structural property of the fusion topology.
New NSF grant on targeted methylation
June 15, 2015
NSF-CBET has funded a joint proposal betweeen our lab and Carl Novina's lab at the Dana-Farber Cancer Institute. The project seeks to develop technology for targeted DNA methylation in human cells.
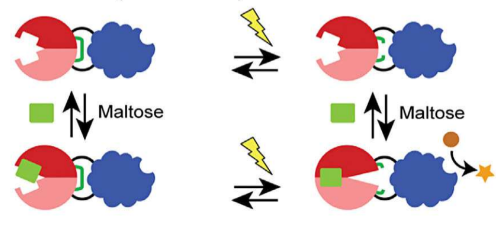
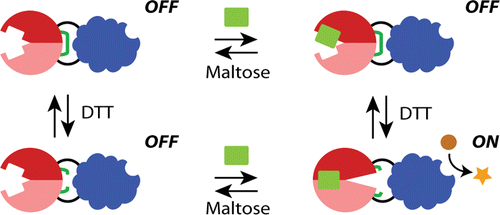
Designing multi-input protein switches based on the conformational ensemble model of allostery
April 22, 2015
Jay and Abby's work on the semi-rational design of multi-input protein switches was published today in Nature Communications. The paper demonstrates how allostery can be established in a non-allosteric fusion protein (between maltose binding protein and TEM1 β-lactamase) through the introduction of mutations designed to increase the conformational entropy of the β-lactamase domain. Biophysical studies support our hypothesis. Our work adds to the growing appreciation of how intrinsically disordered regions can contribute to protein function and lends further support to the conformational ensemble model of allostery. The work was a collaboration with Vince Hilser (JHU).
Improved protein switches with therapeutic potential
November 26, 2014
Clay and Arjun's work on improving our cytosine deaminase switches for use in combination with prodrug therapy is published in PLoS One.
Biotechniques features our targeted methylation research
October 8, 2014
The September issue of Biotechniques has an article on targeting DNA methylation to the genome that features Brian's recent paper in PLoS One.

Jay Choi's paper on protein logic gates published
August 21, 2014
Today ACS Synthetic Biology published Jay Choi's paper on the rational design of protein switches to function as logic gates. The paper describes an alternative approach to gene circuits for introducing logic control elements into cells.
Another new NSF grant
June 17, 2014
More good news from NSF. The lab received a grant from the NSF-DEB Evolutionary Genetics Program to study epistasis.
Brian's paper on improved targeted methyltransferases published
May 8, 2014
Brian's paper on the creation of highly specific DNA methyltransrerases was published today in PLoS One.
New NSF grant on directed evolution
April 21, 2014
The lab received a new grant from the NSF-CBET Biotechnology, Biochemical, and Biomass Engineering Program to explore alternative pathways in directed evolution.
Tiana Warren offered two fellowships
April 18, 2014
Congratulations to Tiana Warren for being selected to receive both an NSF Predoctoral Fellowship and a Ford Foundation Fellowship. Tiana plans to accept the NSF Fellowship.
Marc Ostermeier elected to AIMBE
March 24, 2014
Marc Ostermeier was elected as a Fellow in the American Institute for Medical and Biological Engineering.
Dr. Clay Wright!
March 14, 2014
Congratulations to Clay Wright for successfully defending his thesis today. Next up for Clay (besides getting married) is a postdoc at The University of Washington with Jennifer Nemhauser in the Department of Biology.
Elad's fitness landscape paper published
February 23, 2014
Elad's paper describing a comprehensive map of the mutational landscape of the TEM-1 β-lactamase gene was published online today in Molecular Biology and Evolution.
Dr. Brian Chaikind!
January 17, 2014
Congratulations to Brian Chaikind for successfully defending his thesis today. Next up for Brian is a postdoc at Harvard with David Liu.
Congratulations Dr. Spisak!
October 18, 2024
Congratulations to Shaun Spisak, who successfully defended his PhD thesis "High-thoughput approaches to protein engineering applied to the dCas9 and AAV2 Rep proteins."

Functional and fitness landscapes of E. coli RNase III
February 27, 2023
How protein properties such as protein activity and protein essentiality affect the distribution of fitness effects (DFE) of mutations are important questions in protein evolution. Deep mutational scanning studies typically measure the effects of a comprehensive set of mutations on either protein activity or fitness. Our understanding of the underpinnings of the DFE would be enhanced by a comprehensive study of both for the same gene. Today, Molecular Biology and Evolution published Ryan Weeks' comparative study of the fitness effects and protein activity effects of ∼4,500 missense mutations in the E. coli gene encoding RNase III. RNase III is a global regulator enzyme that cleaves diverse RNA substrates including precursor ribosomal RNA and various mRNAs. He found that although RNase III’s ability to cleave dsRNA is the most important determinant of the fitness effects of rnc mutations, the relationship between RNase III function and cell fitness was complex.Fitness was buffered to small effects on RNase III activity. Differential effects on fitness and functional scores for mutations at highly conserved residues G97, G99, and F188 suggest that these positions may be important for RNase III cleavage specificity.

Genes vary greatly in their propensity for collateral fitness effects of mutations
February 17, 2023
Mutations can have deleterious fitness effects when they decrease protein specific activity or decrease active protein abundance. Mutations will also be deleterious when they cause misfolding or misinteractions that are toxic to the cell (i.e., independent of whether the mutations affect specific activity and abundance). The extent to which protein evolution is shaped by these and other collateral fitness effects is unclear in part because little is known of their frequency and magnitude. Today, Molecular Biology and Evolution published Jacob Mehlhoff's systematic study of the collateral fitness effects in three antibiotic resistance genes: CAT-I, NDM-1, and aadB. He found that genes can have wildly different propensities for collateral fitness effects and that protein aggregation did not correlate with these effects for many of the deleterious mutants in CAT-I and NDM-1. Select deleterious mutantations caused unexpected phenotypes to emerge. The introduction of internal start codons in CAT-1 caused loss of the episome and, even stranger, a mutation in aadB made its cognate antibiotic essential for growth. Jacob's study illustrates how the complexity of the cell provides a rich environment for collateral fitness effects and new phenotypes to emerge.

A dCas9 system for assaying and selecting RNase III activity
December 9, 2022
Pricila Hauk and Ryan Weeks have developed a CRISPR/dCas9 system to assay and select for the ribonuclease activity of RNase III in E. coli. This work was published today in CRISPR J. The key molecule in the system is a hybrid guide RNA (gRNA) between an RNase III substrate and a gRNA that works with dCas9 to repress GFP expression. This fusion must be cleaved by RNase III for full GFP repression. Our system uses GFP fluorescence to report on Escherichia coli RNase III activity in culture and on an individual cell basis, making it effective for selecting individual cells through fluorescence-activated cell sorting. In a forthcoming paper, Ryan Weeks has used the system to determine the in vivo functional effects of thousands of mutations in E. coli RNase III.

A dual selection system for dCas9 activity
June 3, 2022
The nuclease-null CRISPR protein dCas9 (aka CRISPRi) can repress gene expression at sites determined by its cognate guide RNA. The technology would benefit from ways to control this repression exogenously using molecular signals. The creation of such dCas9 switch proteins would benefit from a single system for both positive and negative selection of dCas9 activity. Shaun Spisak has developed such a system, and a paper describing this work was published today in PLOS ONE. This system can be used in E. coli to perform both positive and negative selections for dCas9 activity without additional cloning or transformation steps between selections.

Congratulations Dr. Weeks!
May 20, 2022
Ryan Weeks successfully defended his PhD thesis "Fitness and Functional Landscapes of RNase III."
Nisita Dutta and Olek Pisera receive graduate fellowships
April 13, 2022
Two former Ostermeier lab students recently received prestigious graduate fellowships.
Nisita Dutta (MSE ChemBE 2020) was recently awarded a Gates Cambridge Scholarship, which provides her full funding to pursue a PhD in Chemistry at the University of Cambridge. Nisita will be working with doctors to create nanobody-drug conjugates that target mesothelin, a cell-surface protein expressed on pancreatic cancer cells.
Olek Pisera (BS ChemBE 2017) has been named a 2022 Paul & Daisy Soros Fellow. This fellowship will support his PhD studies in Biomedical Engineering at the University of California, Irvine where he is developing a synthetic biology system that could allow for discovery of antibodies against classes of mammalian cell surface proteins that have been inaccessible with current technologies.
Congratulations Dr. Mehlhoff!
March 14, 2022
Jacob Mehlhoff successfully defended his PhD thesis "Collateral fitness effects of mutations."
Marc Ostermeier elected AAAS Fellow
January 26, 2022
Marc Ostermeier was elected a Fellow of the American Association for the Advancement of Science in the Biological Sciences Section. Read the HUB article on this year's Hopkins' inductees.
New NSF grant to study collateral fitness effects
April 20, 2021
The Ostermeier lab received a ~$1 million grant from NSF-MCB from the Genetic Mechanisms Program to continue our study of collateral fitness effects.
Ostermeier lab certified green thanks to the efforts of Ryan Weeks
February 4, 2021
In December the Ostermeier Lab completed the My Green Lab green lab certification. The program focuses on the behavioral changes that labs can make to reduce their impact on the environment such as setting the -80°C freezer at -70°C, turning off equipment at night, reducing waste, and recycling. The certification represents the global standard for sustainable lab behaviors recognized by the Association for the Advancement of Sustainability in Higher Education, the American Energy Society, and the International Institute for Sustainable Laboratories. Our lab achieved the highest level of certification, green, with over 80% of best lab practices implemented.
The effort was led by PhD student Ryan Weeks, who has also helped launch a pilot program for the university to help 25 Hopkins labs through the certification process as described in a recent HUB article.

Jacob Mehlhoff and Ryan Weeks helping to make N95 masks
May 26, 2020
During the cornovirus research shut down at Hopkins, Jacob Mehlhoff and Ryan Weeks have been volunteering to help make filters for N95 masks at DiPole Materials, an electrospinning company in the Baltimore neighborhood of Pigtown. In total, 13 current students and graduates from the Whiting School of Engineering and Krieger School of Arts and Sciences are now working on the project. By operating 24 hours a day with the help of Hopkins students, the company makes enough material for about 2,000 mask filters per day.
See the story on HUB and the video below.
DiPole Materials: Filter Manufacturing for Medical Masks from Phil DiMuro on Vimeo.
Jacob Mehlhoff receives the WSE Outstanding TA Award
May 13, 2020
Jacob Mehlhoff received the Whiting School of Engineering Award for Outstanding Teaching Assistants for his performance as a TA in Process Design with Aspen.
Congratulations Nisita!
May 9, 2020
Nisita Dutta successfully presented her masters essay "Characterizating the effects of the Psp and Rcs stress pathways on collateral fitness effects in E. coli" This fall Nisita will be starting her MD/PhD with the University of Maryland Medical Scientist Training Program and the NIH Oxford/Cambridge Scholars Program.
Collateral fitness effects of mutations
May 9, 2020
Today, PNAS published our latest work studying the effects of mutations using deep mutational scanning. Mutations provide the source of genetic variability upon which evolution acts. Deleterious protein mutations are commonly thought of in terms of how they compromise the protein’s ability to perform its physiological function. However, mutations might also be deleterious if they cause negative effects on one of the countless other cellular processes. The frequency and magnitude of such collateral fitness effects are unknown. Our systematic study of mutations in a bacterial protein finds widespread collateral fitness effects that were associated with protein aggregation, improper protein processing, incomplete protein transport across membranes, incorrect disulfide-bond formation, induction of stress-response pathways, and unexpected changes in cell properties. The surprising prevalence of deleterious collateral fitness effects suggests they may play a role in constraining protein evolution.
The multi-year effort was led by Jacob Mehlhoff and Frank Stearns with contributions by current and former lab members Dahlia Rohm, Jerry Wang, Henry Tsou, Nisita Dutta, Erin Hsiao, and Courtney Gonzalez with assistance from Alan Rubin at the Walter and Eliza Hall Institute of Medical Research.

Congratulations Jerry!
July 11, 2019
Buheng (Jerry) Wang successfully presented his masters essay "Collateral fitness effects of signal peptide mutants of beta-lactamase."
New proteins for gene-directed enzyme prodrug therapy
June 6, 2019
Two papers were recently published from Tiana Warren's thesis work. Both of the papers involve the yeast enzyme cytosine deaminase (yCD), which together with the prodrug 5-fluorocytosine (5FC) has potential to be used therapeutically in a strategy called Gene-Directed Enzyme Prodrug Therapy (GDEPT). In this strategy, the gene encoding yCD is delivered selectively to cancer cells. Upon subsequent administration of 5FC, the yCD in cancer cells selectively converts 5FC to 5-fluorouracil, a chemotherapeutic. Two limitations of current GDEPT technology are difficulty in selectively delivering the yCD gene to cancer cells and lower than optimal yCD activity in cancer cells.
In a paper appearing in ACS Synthetic Biology, Tiana and her coauthors developed a version of yCD that is selectively degraded in cancer cells because it is fused to a protein domain that is targeted for degradation in hypoxic cells, such as those in tumors. These HOPE fusions have the potential to address the selectivity issue of gene delivery. The second paper appears in AIChE Journal. Tiana and her coworkers used comprehensive site saturation mutagenesis and directed evolution to identify yCD mutations that improve its ability to kill cells in the presence of 5FC. This paper will appear in a special Tribute to Founders issue of AIChE Journal honoring Nobel Laureate Frances Arnold, who was awarded the 2018 Nobel Prize in Chemistry for pioneering the directed evolution of enzymes.
Coauthors of the papers include ChemBE undergraduates Krishna Patel and Jordan Rivera and our JHMI collaborator James Eshleman.

Fitness effects of InDels and sequential mutations in TEM-1
May 9, 2019
The Journal of Molecular Biology recently published two papers by Courtney Gonzalez in which she mapped regions of TEM-1 β-lactamase's fitness landscape using deep mutational scanning
Short insertions and deletions (InDels) are a common type of mutation found in nature and a useful source of variation in protein engineering. InDel events have important consequences in protein evolution, often opening new pathways for adaptation. Yet much less is known about the effects of InDels compared to point mutations and amino acid substitutions. In particular, deep mutational scanning studies on the distribution of fitness effects of mutations have focused almost exclusively on amino acid substitutions. In Courtney's first paper (coauthored by former ChemBE undergradaute student Paul Roberts) she determined the effect of 78% of the 5720 possible amino acid insertions and 98% of the 286 possible amino acid deletions in TEM-1 β-lactamase. This first near-comprehensive examination of the fitness effects of InDels revealed partially overlapping deletion-tolerant and insertion-tolerant regions throughout the protein, especially in unstructured regions and at the end of helices.
In Courtney's second paper she quantified the extent of epistasis between ~8,000 pairs of sequential mutations and found that sequential mutations are particularly prone to interact negatively.

Congratulations Gareth!
April 29, 2019
Gareth Evans successfully presented his masters essay "Creation of modular inducible respressors via directed evolution."
Improved targeted DNA methyltransferases
December 18, 2018
Our latest paper on targeted DNA methylation using CRISPR/dCas9 appeared today in PLoS ONE . We demonstrate how linker length and mutations that reduce the methyltransferase's affinity for DNA can improve targeting of DNA methylation to desired CpG sites. Many thanks to our collaborators in the labs of Winston Timp (Hopkins BME) and Carl Novina (Dana-Farber Cancer Institute).

Congratulations Dr. Gonzalez!
October 5th, 2018
Courtney Gonzalez successfully defended her thesis "Exploring the fitness landscape of TEM-1 β-lactamase: a survey of intragenic epistasis and the fitness effects of InDels."
Congratulations Dr. Warren!
September 28th, 2018
Tiana Warren successfully defended her thesis "Protein engineering improvements to enzyme/prodrug therapy using yeast cytosine deaminase."
Two new NSF grants
June 28, 2018
The Ostermeier lab received two new NSF grants. The aim of the first (from NSF-CBET) is to develop modular protein switches. The aim of the second (from NSF-MCB) is to study underexplored mechanisms that constrain the evolution of proteins.
Congratulations Dr. Xiong!
November 2nd, 2017
Earlier this semester, Tina Xiong successfully defended her thesis on using CRISPR/Cas9 to target DNA methylation of CpG sites. A large portion of her thesis work was published in July in Scientific Reports, with a follow up paper soon to be submitted. Tina is now off to a Senior Scientist position at Merck.
RNA-programmed DNA methylation using CRISPR/Cas9
July 27, 2017
Mammalian genomes exhibit complex patterns of gene expression regulated, in part, by the level of DNA methylation. The advent of engineered DNA methyltransferases (MTases) to target DNA methylation to specific sites in the genome will accelerate many areas of biological research. However, targeted MTases require clear design rules to direct site-specific DNA methylation and minimize the unintended effects of off-target DNA methylation.
Today, Scientific Reports published Tina Xiong's paper describing our use of CRISPR/Cas9 to target methylation. This work is a collaboration between our lab, the Winston Timp lab (JHU), and the Carl Novina lab (Dana-Farber Cancer Institute) including Ostermeier lab alumn Glenna Meister. We describe a targeted MTase composed of an artificially split CpG MTase (sMTase) with one fragment fused to a catalytically-inactive Cas9 (dCas9) that directs the functional assembly of sMTase fragments at the targeted CpG site based on guide RNA sequences. We precisely map RNA-programmed DNA methylation to targeted CpG sites as a function of distance and orientation from the protospacer adjacent motif (PAM). Expression of the dCas9-sMTase in mammalian cells led to predictable and efficient (up to ~70%) DNA methylation at targeted sites. Multiplexing guide RNAs enabled targeting methylation to multiple sites in a single promoter and to multiple sites in multiple promoters. This programmable de novo MTase tool might be used for studying mechanisms of initiation, spreading and inheritance of DNA methylation, and for therapeutic gene silencing.

Tiana Warren receives the 2017 JHU Diversity Recognition Award
May 18, 2017
Congratulations to Tiana Warren for receiving a 2017 JHU Diversity Recognition Award! Tiana has had leadership roles in several organization with the Whiting School of Engineering, most notably the Black Graduate Student Association, the National Organization for the Professional Advancement of Black Chemists and Chemical Engineers, and the Johns Hopkins Diversity Leadership Council. As a KSAS/WSE Graduate Diversity Fellow she has advised divisional leadership on issues related to graduate student diversity and inspired the production of a Homewood-based, graduate URM resource and community handbook.
Elad Firnberg on NPR's Marketplace talking about Revolve
September 30th, 2016
NPR's Marketplace talked to Ostermeier lab alumnus Elad Firnberg yesterday about how free undergrad tuition at Cooper Union enabled him to co-found Revolve Biotechnologies. Revolve Biotechnologies is a life science company focused on engineering better proteins using novel directed evolution technologies. The company's IP portfolio was developed in our lab. In addition to its internal protein pipeline, Revolve is currently offering a service for construction of custom DNA variant libraries.

Congratulations Dr. Shelat!
September 27th, 2016
Earlier this month, Nirav Shelat successfully defended his thesis on engineering the herpes simplex virus thymidine kinase for enzyme prodrug therapy. PLos ONE just published his paper on the development of a new positive genetic selection for nucleoside kinase activity in E. coli.
Tina Xiong wins poster award at SEED!
July 29th, 2016
Congratulations to Tina Xiong for winning a Biotechnology Journal-sponsored poster award at the 2016 Synthetic Biology: Engineering, Evolution & Design (SEED) conference in Chicago last week for her poster entitled "RNA-programmed DNA methylation". Tina earned one of only six poster awards, and there were 234 posters!

Survey of epistasis along an adaptive pathway
June 10th, 2016
An understanding of the role of intragenic epistasis and tradeoffs in adaptation is central to an understanding of protein evolution. Today, Journal of Molecular Biology published Barrett's extensive and systematic study of a series of fitness and epistatic landscapes along an adaptive pathway (Shifting fitness and epistatic landscapes reflect trade-offs along an evolutionary pathway J. Mol. Biol. 428, 2730–2743), the first study of its kind. He measured the effect on ampicillin resistance of ~12,500 unique mutants of alleles of TEM-1 β-lactamase along an adaptive path in the evolution of cefotaxime resistance. This series of fitness and epistatic landscapes provided extensive experimental insight into the relationships between mutation, protein structure, protein stability, and epistasis and revealed the tradeoffs inherent in the evolution of new functions. We found pervasive epistasis involving adaptive mutations that can be partially understood in terms of protein structural and stability considerations. Adaptation moved the protein to a more rugged and precarious region of the fitness landscape, which is a cost of adaptation.

When bad is good
January 29, 2016
Barrett's paper Environmental changes bridge evolutionary valleys (Sci. Adv. 2(1):e1500921) was highlighted in "This Week In Science" in today's issue of Science. The paper demonstrates that evolution through inferior intermediates can paradoxicaly lead to superior outcomes.
Barrett subjected TEM-15 beta-lactamase to four different selection regimen, three of which involved environmental changes that alter the fitness landscape. The "negative selection" regimen (which included steps for selection of mutations that substantially decrease antibiotic resistance) outperformed the other strategies and ultimately resulted in proteins conferring the highest resistance. In the analysis of one such high resistance protein, he found that an initial, severely deleterious mutation is an initial gateway to a relatively inaccessible area of sequence space. Furthermore, this mutation participates in higher-order positive epistasis with a number of beneficial mutations, compensating for the increasing negative epistasis between beneficial mutations as they accumulate (i.e. compensating for diminishing returns). The ability of "negative" selection and environmental changes to provide access to novel fitness peaks has important implications for natural evolutionary mechanisms as well as applied directed evolution.

Modular protein switches built from antibody mimetics
December 7, 2015
In a just published paper in Protein Engineering Design and Selection, we demonstrate that protein switches can be built using DARPins and monobodies as input domains. The DTRA and NIH funded research was initiated by Manu Kanwar and Amol Date and fully realized by first author Nate Nicholes (with some assistance from Pamphile Beaujean and Pricila Hauk).
Our manuscript describes how domain insertion can be used to create protein switches that consist of fusions between antibody mimetic proteins (monobodies and DARPins) and the enzyme beta-lactamase. These switches' enzyme activity is modulated by the antibody mimetic domain’s ligand. We demonstrate that these switches exhibit modularity in the sense that new switches can be created simply through the introduction of mutations in the antibody mimetic domain that are known to cause binding to the new ligand. There are two significant conclusions from our work. First, it is possible to create protein switches by domain insertion using input domains that do not undergo large conformational changes upon ligand binding. Second, DAPRins and, to a lesser extent, monobodies show promise as input domains for a modular platform for the rapid development of switches.

Two recent protein switch papers
October 13, 2015
Two of our papers on protein switches have recently appeared in Biotechnology and Bioengineering.
In the first paper, Jay Choi together with Maya Zayats from Peter Searson's lab show how an electrochemical signal can be used as an exogenous input to control protein switch function via reduction of the engineered disulfide bonds. This study suggests that a disulfide-containing protein switch is a potentially useful platform for bioelectronic sensors with remote control of the sensing ability.
In the second paper, Nate Nicholes and Jen Tullman led an effort to create switches by fusing TEM-1 beta-lactamase and a variety of paralogous periplasmic binding proteins. The results indicate that the emergence of the switch property likely depends on the precise molecular details of the fusions and cannot be easily predicted from some overall general structural property of the fusion topology.

New NSF grant on targeted methylation
June 15, 2015
NSF-CBET has funded a joint proposal betweeen our lab and Carl Novina's lab at the Dana-Farber Cancer Institute. The project seeks to develop technology for targeted DNA methylation in human cells.
Designing multi-input protein switches based on the conformational ensemble model of allostery
April 22, 2015
Jay and Abby's work on the semi-rational design of multi-input protein switches was published today in Nature Communications. The paper demonstrates how allostery can be established in a non-allosteric fusion protein (between maltose binding protein and TEM1 β-lactamase) through the introduction of mutations designed to increase the conformational entropy of the β-lactamase domain. Biophysical studies support our hypothesis. Our work adds to the growing appreciation of how intrinsically disordered regions can contribute to protein function and lends further support to the conformational ensemble model of allostery. The work was a collaboration with Vince Hilser (JHU).

Improved protein switches with therapeutic potential
November 26, 2014
Clay and Arjun's work on improving our cytosine deaminase switches for use in combination with prodrug therapy is published in PLoS One.
Biotechniques features our targeted methylation research
October 8, 2014
The September issue of Biotechniques has an article on targeting DNA methylation to the genome that features Brian's recent paper in PLoS One.

Jay Choi's paper on protein logic gates published
August 21, 2014
Today ACS Synthetic Biology published Jay Choi's paper on the rational design of protein switches to function as logic gates. The paper describes an alternative approach to gene circuits for introducing logic control elements into cells.

Another new NSF grant
June 17, 2014
More good news from NSF. The lab received a grant from the NSF-DEB Evolutionary Genetics Program to study epistasis.
Brian's paper on improved targeted methyltransferases published
May 8, 2014
Brian's paper on the creation of highly specific DNA methyltransrerases was published today in PLoS One.
New NSF grant on directed evolution
April 21, 2014
The lab received a new grant from the NSF-CBET Biotechnology, Biochemical, and Biomass Engineering Program to explore alternative pathways in directed evolution.
Tiana Warren offered two fellowships
April 18, 2014
Congratulations to Tiana Warren for being selected to receive both an NSF Predoctoral Fellowship and a Ford Foundation Fellowship. Tiana plans to accept the NSF Fellowship.
Marc Ostermeier elected to AIMBE
March 24, 2014
Marc Ostermeier was elected as a Fellow in the American Institute for Medical and Biological Engineering.
Dr. Clay Wright!
March 14, 2014
Congratulations to Clay Wright for successfully defending his thesis today. Next up for Clay (besides getting married) is a postdoc at The University of Washington with Jennifer Nemhauser in the Department of Biology.
Elad's fitness landscape paper published
February 23, 2014
Elad's paper describing a comprehensive map of the mutational landscape of the TEM-1 β-lactamase gene was published online today in Molecular Biology and Evolution.
Dr. Brian Chaikind!
January 17, 2014
Congratulations to Brian Chaikind for successfully defending his thesis today. Next up for Brian is a postdoc at Harvard with David Liu.
LATEST PUBLICATIONS
R. Weeks, M. Ostermeier
Fitness and functional landscapes of the E. coli RNase III gene rnc
Mol. Bio. Evol. 40, msad047 (2023) [full text]
J. D. Mehlhoff, M. Ostermeier
Genes vary greatly in their propensity for collateral fitness effects of mutations
Mol. Bio. Evol. 40, msad038 (2023) [full text]
P. Hauk, R. Weeks, M. Ostermeier
A CRISPR/dCas9 system for assaying and selecting for RNase III activity in vivo in E. coli
CRISPR J. 6, 43-51 (2023) [full text]
S. Spisak, B. O'Brien, M. Ostermeier
A bacterial dual positive and negative selection system for dCas9 activity
PLoS One 17(6), e0269270 (2022) [full text]
S. Spisak, M. Ostermeier
Engineered protein switches for exogenous control of gene expression
Biochem. Soc. Trans. 48, 2205-2212 (2020) [full text]
J. D. Mehlhoff, F. W. Stearns, D. Rohm, B. Wang, E.-Y. Tsou, N. Dutta, M.-H. Hsiao, C. E. Gonzalez, A. F. Rubin, M. Ostermeier
Collateral fitness effects of mutations
Proc. Nat. Acad. Sci. USA 117, 11597-11607 (2020) [full text] [BioRxiv]
J. D. Mehlhoff, M. Ostermeier
Biological fitness landscapes by deep mutational scanning
Methods Enzymol. 643, 203-224 (2020) [full text]
T. D. Warren, K. Patel, J. L. Rivera, J. R. Eshleman, M. Ostermeier
Comprehensive mutagenesis on yeast cytosine deaminase yields improvements in 5-fluorocytosine toxicity in HT1080 cells
AIChE Journal, 66:e16688 (2020) [full text]
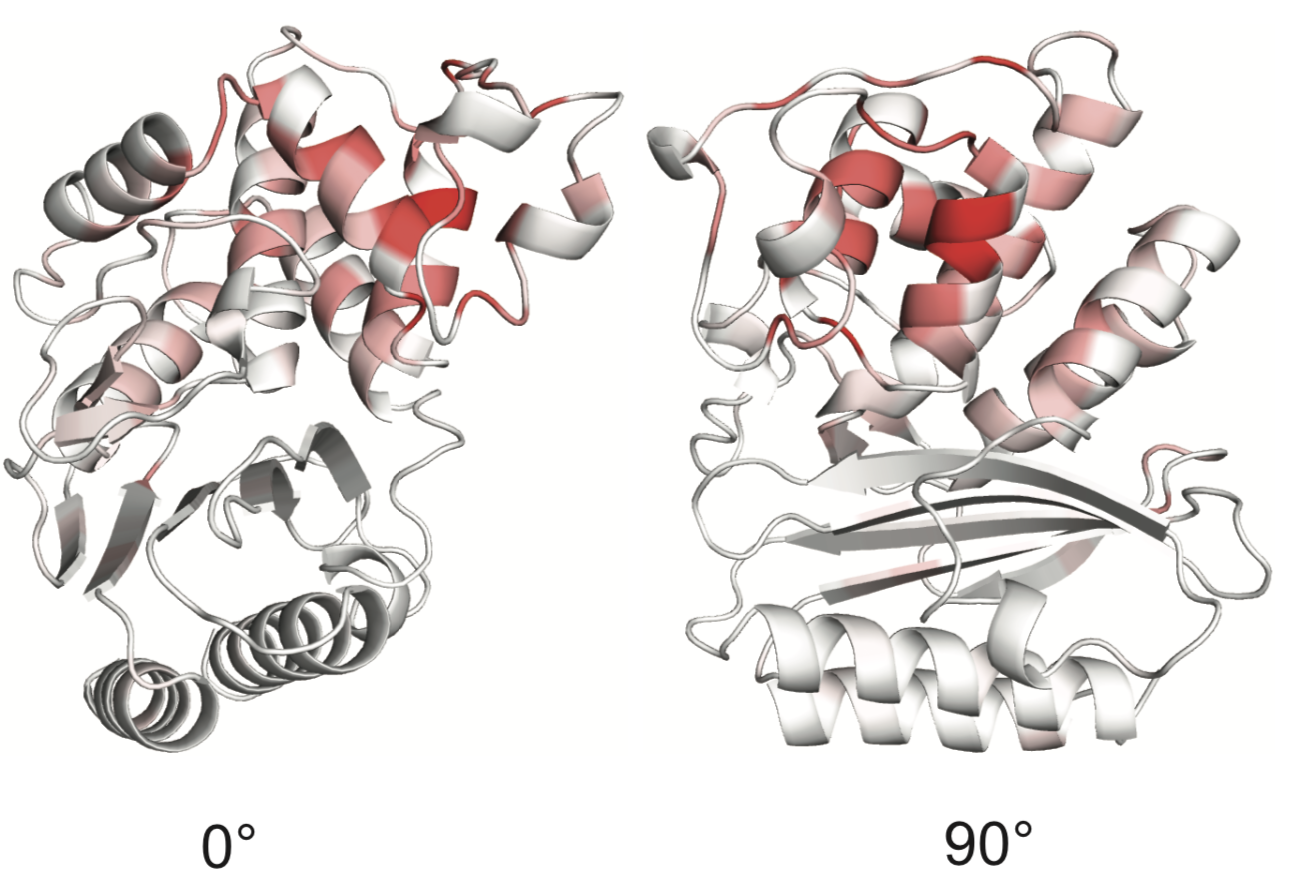
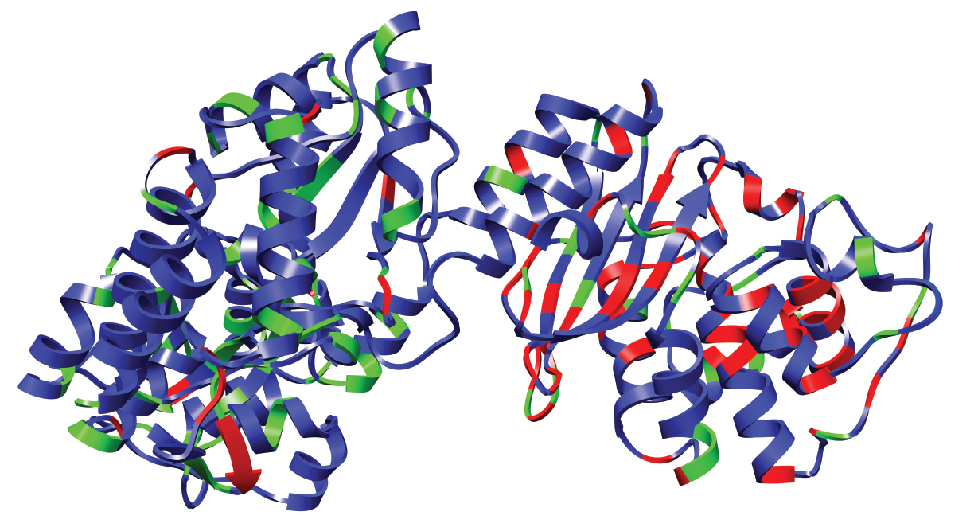
Sites of collateral fitness effects in TEM-1 β-lactamase.
R. Weeks, M. Ostermeier
Fitness and functional landscapes of the E. coli RNase III gene rnc
Mol. Bio. Evol. 40, msad047 (2023) [full text]
J. D. Mehlhoff, M. Ostermeier
Genes vary greatly in their propensity for collateral fitness effects of mutations
Mol. Bio. Evol. 40, msad038 (2023) [full text]
P. Hauk, R. Weeks, M. Ostermeier
A CRISPR/dCas9 system for assaying and selecting for RNase III activity in vivo in E. coli
CRISPR J. 6, 43-51 (2023) [full text]
S. Spisak, B. O'Brien, M. Ostermeier
A bacterial dual positive and negative selection system for dCas9 activity
PLoS One 17(6), e0269270 (2022) [full text]
S. Spisak, M. Ostermeier
Engineered protein switches for exogenous control of gene expression
Biochem. Soc. Trans. 48, 2205-2212 (2020) [full text]
J. D. Mehlhoff, F. W. Stearns, D. Rohm, B. Wang, E.-Y. Tsou, N. Dutta, M.-H. Hsiao, C. E. Gonzalez, A. F. Rubin, M. Ostermeier
Collateral fitness effects of mutations
Proc. Nat. Acad. Sci. USA 117, 11597-11607 (2020) [full text] [BioRxiv]
J. D. Mehlhoff, M. Ostermeier
Biological fitness landscapes by deep mutational scanning
Methods Enzymol. 643, 203-224 (2020) [full text]
T. D. Warren, K. Patel, J. L. Rivera, J. R. Eshleman, M. Ostermeier
Comprehensive mutagenesis on yeast cytosine deaminase yields improvements in 5-fluorocytosine toxicity in HT1080 cells
AIChE Journal, 66:e16688 (2020) [full text]

Sites of collateral fitness effects in TEM-1 β-lactamase.
