MAJOR RESEARCH AREAS
last updated June 7th, 2019
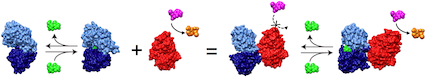
Protein switches
funded by NSF-CBET and Pebble Labs
The incredible complexity of biological systems derives from the high degree of interactions amongst their constituent components. As such, the cell is often described as a complex circuit consisting of an interacting network of molecules. A key component of these networks are protein switches that serve to couple cellular functions. A switch changes its activity state (the output) in the presence of a biomolecular signal (the input). Examples of natural switches include allosteric enzymes which couple effector levels to enzymatic activity and ligand-dependent transcription factors that couple ligand concentration to gene expression. The ability to create switches comprised of any desired input and output functions would enable the rewiring of the cellular circuitry to our own design and has tremendous potential for developing novel molecular sensors, selective protein therapeutics, and as a tool for elucidating molecular and cellular function.
Our directed evolution strategy for switch construction involving the recombination of genes encoding the prerequisite input and output functions for the switch has proven very successful. We have created ligand-activated enzymes with up to 600-fold changes in enzyme activity in response to the presence of a ligand and developed switches in which a cancer marker triggers prodrug activation. Our current research efforts are divided between seeking an understanding of switch mechanisms and applying our techniques to create switches for applications.
Protein evolution
funded by NSF-MCB
The fitness landscape model, as first conceptualized by John Maynard Smith in 1970, is a landmark concept for the fields of molecular evolution and macromolecular structure and function. The model imagines evolution as a process by which a sequence moves by mutations across the fitness landscape. The nature of the fitness landscape fundamentally shapes evolution. Using TEM-1 β-lactamase as a model gene, we are building a comprehensive and detailed maps of the fitness effects of mutations. These experiments have brought insight into the determinants of protein mutational effects, the interactions of mutations, the origins of the genetic code, the determinants of protein evolution rates, and natural evolutionary mechanisms. In addition we are using these results to formulate new approaches for directed evolution.
last updated June 7th, 2019
Protein switches
funded by NSF-CBET and Pebble Labs
The incredible complexity of biological systems derives from the high degree of interactions amongst their constituent components. As such, the cell is often described as a complex circuit consisting of an interacting network of molecules. A key component of these networks are protein switches that serve to couple cellular functions. A switch changes its activity state (the output) in the presence of a biomolecular signal (the input). Examples of natural switches include allosteric enzymes which couple effector levels to enzymatic activity and ligand-dependent transcription factors that couple ligand concentration to gene expression. The ability to create switches comprised of any desired input and output functions would enable the rewiring of the cellular circuitry to our own design and has tremendous potential for developing novel molecular sensors, selective protein therapeutics, and as a tool for elucidating molecular and cellular function.
Our directed evolution strategy for switch construction involving the recombination of genes encoding the prerequisite input and output functions for the switch has proven very successful. We have created ligand-activated enzymes with up to 600-fold changes in enzyme activity in response to the presence of a ligand and developed switches in which a cancer marker triggers prodrug activation. Our current research efforts are divided between seeking an understanding of switch mechanisms and applying our techniques to create switches for applications.

Protein evolution
funded by NSF-MCB
The fitness landscape model, as first conceptualized by John Maynard Smith in 1970, is a landmark concept for the fields of molecular evolution and macromolecular structure and function. The model imagines evolution as a process by which a sequence moves by mutations across the fitness landscape. The nature of the fitness landscape fundamentally shapes evolution. Using TEM-1 β-lactamase as a model gene, we are building a comprehensive and detailed maps of the fitness effects of mutations. These experiments have brought insight into the determinants of protein mutational effects, the interactions of mutations, the origins of the genetic code, the determinants of protein evolution rates, and natural evolutionary mechanisms. In addition we are using these results to formulate new approaches for directed evolution.
FUNDING
Our research is currently funded by NSF-MCB, NSF-CBET, and Pebble Labs.
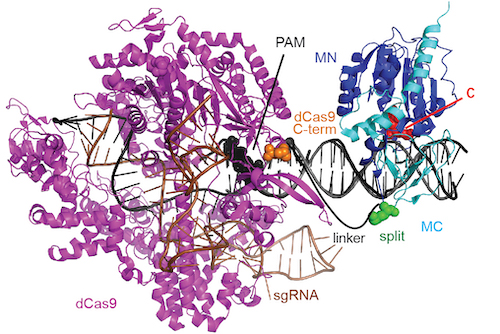
Model of a dCas9-directed cytosine methyltransferase
Our research is currently funded by NSF-MCB, NSF-CBET, and Pebble Labs.

Model of a dCas9-directed cytosine methyltransferase
